Elucidating the Architecture of Plant Gene Regulatory Networks
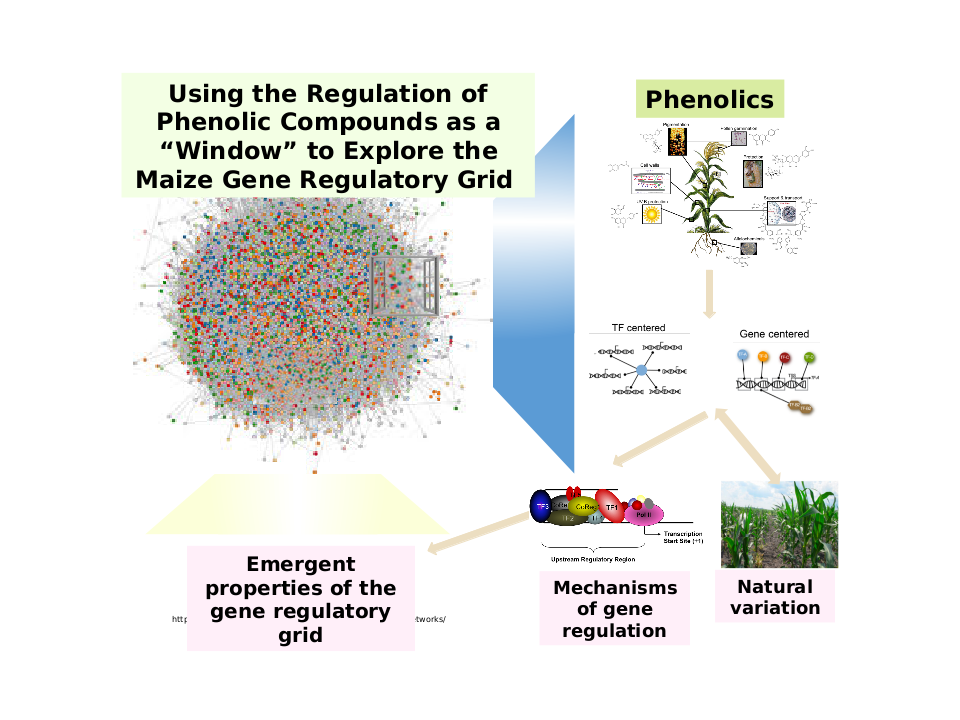
Figure 1. Using the Regulation of Phenolic Compounds as a Window for Exploration
into the Maize Gene Regulatory Network. The integration of multiple omics data related to phenolic metabolism helps reveal the
properties of the gene regulatory grid in maize. Focusing on all different types of
phenomics, genomics, transcriptomics, and metabolomics data allow us to study the
complex inter-relationships and how they function toward influencing phenotypes, such
as phenolic compounds accumulation in maize population. With the rapid expansion within
the omics field, consolidating multiple approaches are essential to provide new perspectives
on how the gene regulatory grid/network makeup of maize.
Elucidating the architecture of gene regulatory grids and gene regulatory networks
involves linking the cis-regulatory machinery (cistrome) with the trans-acting factors
through the identification of protein-DNA interactions (PDIs). Our lab combines gene-
and transcription factor (TF)-centered approaches to determine the biologically-relevant
PDIs important for the temporal and spatial expression of all genes. Experiments are
largely conducted in maize and Arabidopsis as well as projects in tomato and Camelina. In
tomato, we are investigating the interaction of several TFs with target genes involved
in early fruit development. In maize, we implemented DNA affinity purification sequencing (DAP-seq) and chromatin
immunoprecipitation sequencing (ChIP-seq) to establish the genome-wide occupancies
of 45 TFs involved in the control of the phenylpropanoid pathway. The cistrome hinges
on where transcription initiation starts, thus, our lab has been conducting extensive
cap analysis of gene expression (CAGE) to determine the location of transcription
start sites (TSS) in two maize inbred lines, B73 and Mo17, under normal and stress
conditions. Additionally, we aim to understand how TSS selection is controlled in
cis/trans; thus, we have conducted CAGE sequencing and computational analysis on F1
hybrids to generate unparalleled information on how TSSs are determined.
Biosynthesis, Transport and Regulation of Flavonoids and Other Phenolic Compounds
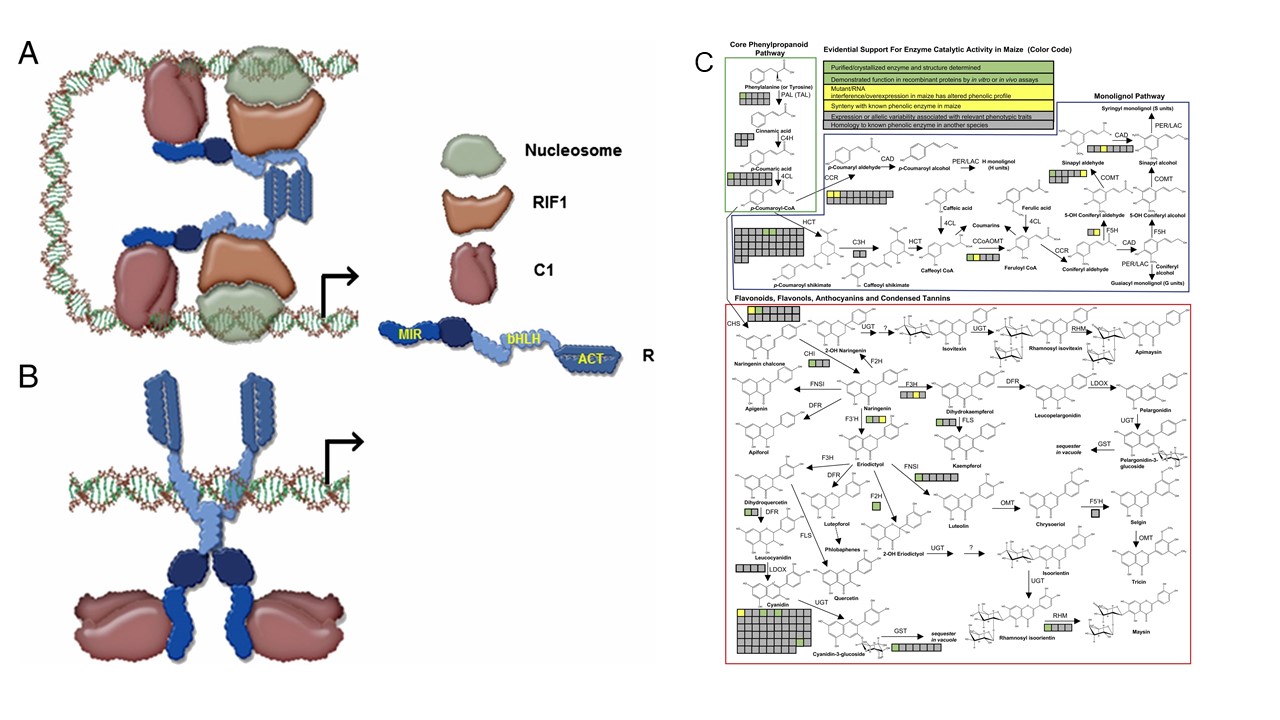
Figure 2. Biosynthesis, Transport and Regulation of Flavonoids and Other Phenolic
Compounds. Proposed model for promoter switching by R (bHLH protein) to regulate anthocyanin
accumulation in association with C1 (R2R3-MYB protein; 2A) and RIF1 (EMSY-related
protein) in Maize (Kong et al 2012; 2B). Summary of evidence for Core Phenylpropanoid,
Monolignol, and Flavonoid Pathways in Maize (Gomez-Cano et al 2020; 2C).
Phenolic compounds are important plant specialized metabolites that participate in
biotic and abiotic interactions between plants and the environment. Especially, Flavonoid-pigments,
including the anthocyanins and phlobaphenes, have provided powerful visual tools to
understand fundamental biological concepts. Our lab is interested to investigate how
the interaction between R2R3-MYB and basic helix-loop-helix (bHLH) transcription factors
cooperate to regulate anthocyanin accumulation in maize and Arabidopsis; and how bHLH transcription
factors with very similar DNA-binding domains display regulatory specificity, for
example in the regulation of flavonoid pigments in maize aleurone, and in Arabidopsis epidermal
cell differentiation (e.g., trichomes). Meanwhile, our lab is using anthocyanin compounds
to better understand how specialized metabolites traffick within cells, an issue for
the vast majority of thousands of specialized metabolites that characterizes land
plants. Given the importance of phenylpropanoids and lignin compounds for agricultural
biomass and biofuel potential, our lab is also focused on the identification of new genes
involved in the biosynthesis and regulation of maize phenylpropanoids, using a combination
of yeast and transient expression in Nicotiana benthamiana.
Engineering the Pathways for Insecticidal and Nutritional Flavones
.png)
Figure 3. Engineering the Pathways for Insecticidal and Nutritional Flavones. A transgenic tomato plant harboring the gene, ZmFNSI-1, driven by the pMKS1 trichome
specific promoter, encodes a flavone synthase, which uses the flavonones naringenin
and eriodyctiol to produce the flavones apigenin and luteolin, respectively (A). By
expressing ZmFNSI-1 in a tissue specific manner, we aim to obtain flavones beneficial
to human health and precursors (luteolin) in maysin biosynthesis, our main target
to engineer in tomato. To engineer flavones and C-glycosylflavones involved in the
maysin biosinthesis, all the enzymes are being expressed in a tissue specific manner
using the pMKS1 promoter (B) and in constitutive manner using the promoter of the
Chalcone synthase gene, pCHI1 (C).
One group of flavonoid compounds with human health importance and that also provide
resistance to herbivores and other pathogens are flavones. In maize, C-glycosylflavones,
in particular maysin, provide significant protection to the corn earworm (Helicoverpa zea)
and related insects when they accumulate in the silks. We have elucidated the pathway
for maysin biosynthesis, and we are currently engineering this pathway into other
crops and vegetables that are susceptible to Helicoverpa zea. Using stable transformation
in tomato and transient expression in Nicotiana benthamiana, we are expressing the
necessary enzyme-coding genes to produce all the flavones and C-glycosyl flavones
involved in maysin biosynthesis. Because glycosylated flavones are usually less health-beneficial
to animals than the aglycones, we are also combining available maize mutants to enhance
the accumulation of seed flavone aglycones, in particular apigenin and luteolin.
Biosynthesis and Regulation of Seed Oils in Emerging Oilseed Crops

As part of several collaborative projects, the lab is involved in elucidating the
mechanisms by which seed oils are regulated in Camelina and Pennycress, and how this
knowledge can be translated into strategies to enhance oilseed production in these
plants with increasing agronomic potential. We are currently working on transforming
candidate genes responsible for enhancing or regulating seed oil production in Camelina and
Pennycress. Transgenes altering oil accumulation allow us to increase fundamental
knowledge of the not fully understood fatty acid biosynthetic pathways, as well as, enable targeted
breeding to increase the yield of oil in the seed. In addition, we also plan to accomplish
this goal with DNA affinity purification sequencing (DAP-seq) strategy, an in vitro TF
centered technique, to identify potential TF-DNA interactions responsible for elevating
oil levels.
Image courtesy of: CINTIA ARIAS of University of North Texas, Texas (UNT)